This is a guest post written with Michael Shamash, a Master’s student in the Maurice Lab at Canada's McGill University where he currently studies childhood malnutrition and the active role of bacteriophages (viruses that can only infect bacteria) in the gut microbiome.
In microbiology, a plaque is defined as a “clear area on an otherwise opaque field of bacteria that indicates the inhibition or dissolution of the bacterial cells by some agent, either a virus or an antibiotic. [Plaques] are a sensitive laboratory indicator of the presence of some anti-bacterial factor.”
I had the good fortune of meeting Michael (virtually) in June 2021. He told me a story of a young girl that had suffered from an antibiotic-resistant infection and how bacteriophages, viruses that only infect bacteria, were ultimately able to successfully treat the infection (original Nature Medicine article here). Michael sought to help streamline some of the monotonous aspects of studying and preparing bacteriophages by harnessing the power of computer vision and AI. Using Roboflow’s annotation and image processing tools, Michael was able to go from an idea to a fully functional iOS app within only 5 weeks!
Written by Michael Shamash
The Problem
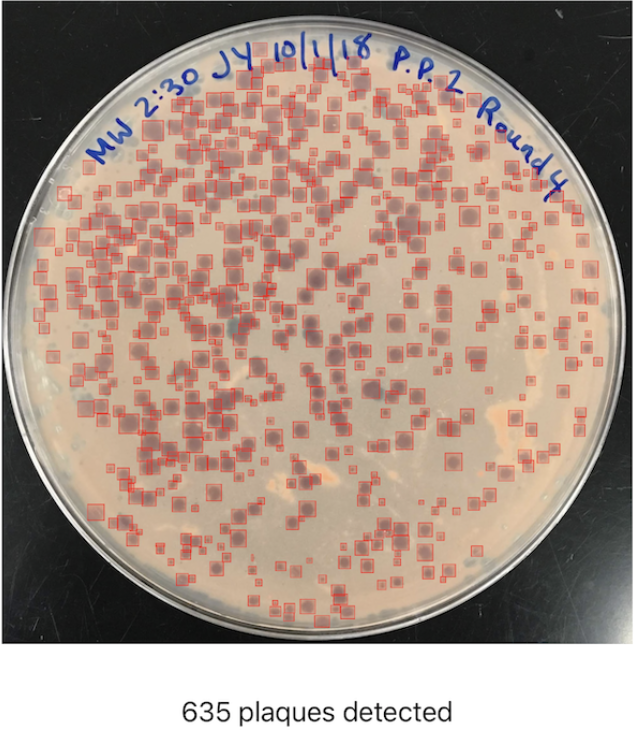
When working with bacteriophages (phages), scientists often need to perform the time-intensive monotonous task of counting plaques on Petri dishes. To help solve this problem I developed OnePetri, a set of machine learning models and a mobile phone application (currently iOS-only) that accelerates common microbiological Petri dish assays using AI.
A task that once took microbiologists several minutes to do per Petri dish (adds up quickly considering there are often tens of Petri dishes to analyze at a time!) could now be mostly automated thanks to computer vision, and completed in a matter of seconds...don’t blink or you’ll miss it!
For more information and to download OnePetri please visit https://onepetri.ai/.
The dataset is also available on Roboflow Universe.
Announcement for the release of OnePetri (iOS) and a short video of the app in action. By @michaelviridae
Using Roboflow to annotate, pre-process, and augment training datasets
Under the hood, OnePetri sports two YOLOv5s object detection models for Petri dish and plaque identification. These models were trained on a diverse set of images from the HHMI's SEA-PHAGES program. I’m extremely grateful for their contribution to this project, as this wouldn't have been possible without their support!
A total of 43 images were included in the initial dataset for OnePetri’s plaque detection model (v1.0) with the following split: 29 training, 9 validation, 5 test. Although the number of images is quite small, each has a large number of plaques (~100 plaques per image on average), giving the algorithm plenty of data to train on.
The following pre-processing steps were applied: auto-orient, tile image into 5 rows x 5 columns, resize tiles to 416px x 416px. To increase the training dataset even further, the following augmentation steps were applied: grayscale (35% of images), hue shift (-45° to +45°), blur up to 2px, mosaic.
Within only two weeks, I managed to fully annotate the initial dataset and train the first set of models!
From model to market: bringing OnePetri to the iOS App Store
Initially, I began training and testing the various YOLOv5s models on the Google Colab platform. Given that this is a free resource and is shared by others, the available GPU resources can be lacking at times. I decided to continue training locally using an NVIDIA RTX 2060 Super GPU to speed things up and increase the total VRAM I’d have available for use.
Once trained, the models were converted to a format that would be compatible with Apple’s CoreML APIs, and so began the process of building OnePetri for iOS! A few weeks later I had a functional prototype which was put through its paces by a dedicated group of beta testers who work with phage regularly. The OnePetri Swift source code is now publicly available on GitHub under a GPL-3.0 license. The trained models are also publicly available on GitHub.
Upon opening OnePetri, the user is prompted to select or take a photo for analysis, which can contain any amount of Petri dishes for analysis. Once a Petri dish is selected, the cropped Petri dish image is tiled into overlapping 416px x 416px squares, and plaques are inferred serially on each tile. After plaque de-duplication, the final result is presented to the user with a summary of the total number of plaques detected and bounding boxes around each plaque.
All image processing and inference is done locally on-device, meaning OnePetri can be used even without internet access in remote locations once the app has been installed.
I’m happy to say that OnePetri is now available to download for free on the Apple App Store!
What’s next for OnePetri?
Going forward, I’ll continue to iterate and improve upon the models included in OnePetri by incorporating user-submitted images, as well as additional images from the HHMI SEA-PHAGES program, into the training datasets. In addition to plaque counts, I’m working on adding support for bacterial colony counting and other assays. Finally, I’m also working on bringing OnePetri to a wider range of platforms, increasing access to this time-saving tool to all phage scientists across the globe - so stay tuned!
If you’d like to learn more about Michael, his work on OnePetri, or his current research, below are some links to explore.
Michael's Links: Personal Site | LinkedIn | Twitter | Publications: Google Scholar, ORCiD | GitHub


